
Transforming Medical Laboratory Science with Vision-Language Models: A Focus on Microscopy in Microbiology, Hematology, and Histopathology
ABSTRACT
KEYWORDS
HTML
Introduction
Microscopy has historically been fundamental in medical laboratory diagnostics, providing direct visualization of cellular and microbial structures crucial for the identification of infectious, haematologic, and histopathological diseases. In microbiology, techniques such as Gram staining, Ziehl–Neelsen staining, and wet mounts offer swift initial assessments of bacterial, fungal, and parasitic infections. In hematology, the analysis of peripheral blood smears facilitates a comprehensive evaluation of erythrocyte and leukocyte morphology, assisting in the identification of anemia, hematologic malignancies, and platelet abnormalities. The histopathological assessment of stained tissue sections, especially with hematoxylin and eosin, aids in the analysis of architectural and cytological characteristics pertinent to malignancy, inflammation, and tissue degeneration. These routines are crucial to clinical decision-making, disease surveillance, and medical education [1].
Notwithstanding its diagnostic significance, conventional microscopy is limited by its manual characteristics and reliance on human proficiency. Variability in interpretation, operator tiredness, and training discrepancies contribute to diagnostic errors and diminished reproducibility. In high-throughput or resource-constrained environments, the scarcity of competent workers, protracted slide evaluations, and infrastructural inadequacies impede prompt and precise diagnosis. These issues show how important it is to have systems that can be scaled up, copied, and improved using computers to help people make better decisions and reduce workload bottlenecks.
Artificial intelligence (AI), especially deep learning approaches like convolutional neural networks (CNNs), has been useful in automating the process of finding patterns in medical images in domains like radiology, pathology, and cytology. However, CNNs usually give outputs that can't be understood, like classification scores or bounding boxes, because they can't put results in terms that are useful for therapy. Because of this limitation, there has been more interest in multimodal models that include an understanding of both text and images [2].
Vision-Language Models (VLMs) are a new type of AI that makes it possible to understand both images and words at the same time. Utilizing architectures like CLIP (Contrastive Language–Image Pretraining) and GPT-style language models, VLMs can produce structured descriptions or diagnostic narratives from visual input, so aligning more closely with the interpretive processes of laboratory specialists. A VLM can classify a Gram-stained image while simultaneously detailing cell morphology, staining characteristics, and probable organism classification—offering more comprehensive contextual information than unimodal models [3].
This article critically analyses the function of VLMs in enhancing diagnostic processes within three areas of medical microscopy: microbiology, hematology, and histopathology. We consolidate contemporary evidence from peer-reviewed research to evaluate the capabilities, performance measures (e.g., accuracy, AUC, F1 scores), and limits of current VLMs in these domains. Additionally, we tackle critical technological, ethical, and implementation difficulties, including data annotation quality, generalisability across varied clinical environments, privacy concerns, and regulatory monitoring. This review seeks to establish a practical and evidence-based framework for the incorporation of VLMs into clinical laboratory practice by contextualizing them within the larger scope of diagnostic automation. To elucidate the interpretative disparity between classic AI models and Vision-Language Models (VLMs), we present a visual example (Figure 1) that illustrates how each model type examines and conveys results from the identical haematologic smear. This contrast highlights the interpretability advantage of VLMs in generating elaborate, human-like narratives [4].
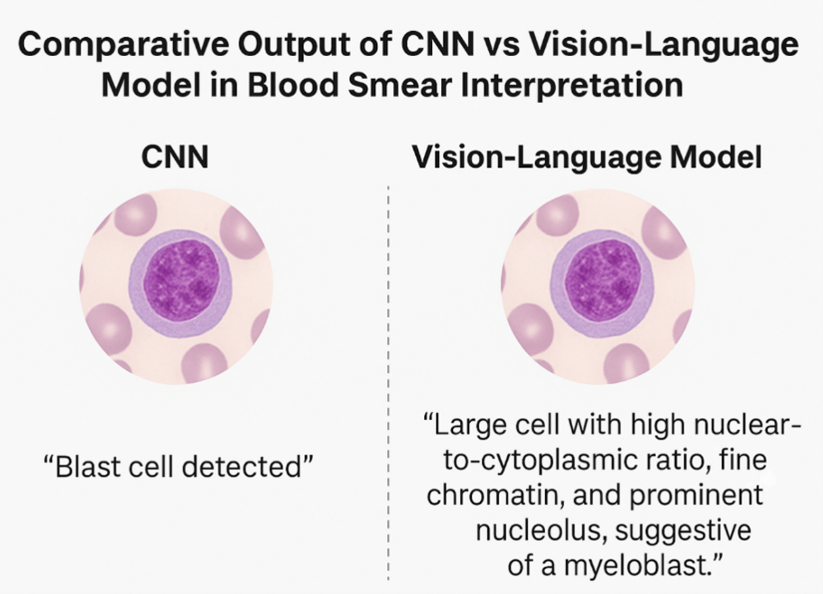
This graphic displays a comparative analysis of outputs generated by a Convolutional Neural Network (CNN) and a Vision-Language Model (VLM) reading the same blood smear image. The CNN assigns a categorical label—“Blast cell detected”—whereas the VLM generates a comprehensible descriptive report: “Large cell with a high nuclear-to-cytoplasmic ratio, fine chromatin, and prominent nucleolus, indicative of a myeloblast.” This comparison underscores the narrative interpretability of VLMs, which emulate human diagnostic reasoning more proficiently than conventional CNN outputs.
Overview of Vision-Language Models (VLMS)
Vision-Language Models (VLMs) are a type of multimodal AI system that can interpret and combine visual and written input at the same time. Unlike traditional unimodal models that only work with images or text, VLMs combine these two types of data. This makes it easier to do things like captioning photos, answering visual questions, and reasoning across multiple modalities. These qualities make it easier to move from static categorization to outputs that are easier to understand. This lets models describe visual content in coherent natural language and connect it to contextual information.
Many different VLM designs work well on general-purpose datasets. Some of these models combine visual features with semantic text embeddings, while others add visual inputs to language models. In medical imaging, these features make it possible to find morphological patterns and make descriptive narratives that match the clinical record. For instance, when used on annotated pathology photos, VLMs can find features like cellular atypia or structural disorganization and present the results in structured prose that seems like a report from an expert [5].
This is different from standard convolutional neural networks (CNNs), which usually just give one output label without any context. Convolutional Neural Networks (CNNs) can correctly sort cell kinds or disease classifications, but they can't explain why they make the predictions they do. On the other hand, VLMs can give you a lot of information, including seeing Gram-positive cocci in chains and suggesting how to group them, or finding macrocytic erythrocytes next to hypersegmented neutrophils, which are signs of certain blood illnesses. These narrative outputs help with diagnosis and serve instructional and documentation purposes.
VLMs help with both pattern recognition and verbal expression of findings in microscopy-based diagnostics, which include microbiology, hematology, and histopathology. These are both important for laboratory reporting. Their outputs can help lab scientists by giving them a first look at things that need to be evaluated and validated, which could lower the number of different diagnoses and the time it takes to get results. Also, in places where resources are limited and there aren't enough trained specialists, VLMs may help with diagnosis by standardizing outputs and finding unusual patterns for experts to look at [6].
There are usually four steps to using VLMs in microscopy workflows: getting high-resolution digital images, processing them by the VLM to make structured descriptions, connecting with laboratory information systems for documentation, and having experts check the results before reporting. This method makes it easier to create a hybrid framework for AI-assisted diagnostics that puts a premium on transparency and reproducibility while having human oversight [5].
There are several possible benefits to using VLMs in microscopy, but their usage must be controlled by strict validation, clear performance standards, and appropriate regulatory frameworks. They are a useful tool for improving diagnostic procedures because they can produce standardized, understandable outputs. However, their real effectiveness depends on how well the model holds up, how good the training data is, and how well it works with current laboratory equipment. There are four steps to adding VLMs to microscopy workflows: getting high-resolution digital images, having the VLM automatically interpret them, making structured diagnostic descriptions, and having laboratory professionals check them. Figure 2 shows an example of this technique, which is a hybrid kind of human–AI collaboration in which automation helps but does not replace expert review. The goal of this system is to make it easier to get diagnostic services in both resource-rich and resource-limited settings, as well as to make them easier to repeat and shorten the time it takes to report [7].
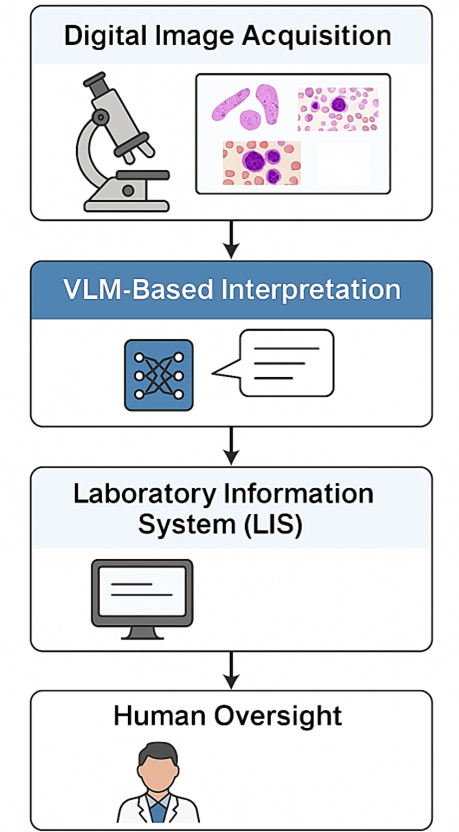
Utilization of Vision-Language Models in Microscopy
The application of Vision-Language Models (VLMs) in microscopy signifies a significant advancement in computational pathology, especially in the automation of intricate visual interpretation tasks that have historically relied on skilled human assessment. By integrating visual and textual modalities, VLMs create novel avenues for scalable and interpretable diagnostics. Nonetheless, their implementation necessitates a careful assessment of their practical accuracy, repeatability, and contextual constraints within laboratory settings. This section offers a specialized evaluation of VLM integration in microbiology, hematology, and histopathology.
Microbiology
Microscopy is essential for the prompt detection of infectious pathogens by techniques such as Gram stain and culture analysis. These procedures, although swift and informative, necessitate a sophisticated assessment of morphology, staining quality, and spatial arrangement—abilities that frequently differ based on experience and institutional training.
Clinical Workflow Example: Gram-stain triage (pre-/analytical/post-analytical).
Pre-analytical: After slide preparation and staining, the system ingests a low-magnification overview plus 2–3 high-power fields; specimen metadata (source, ward, recent antibiotics) is pulled from the LIS. Analytical: The VLM screens for “organisms present/absent,” proposes Gram category and basic morphology (e.g., “Gram-positive cocci in clusters”), and produces a short rationale grounded in highlighted regions. Targets: Site-defined goals include ≥95% sensitivity for “organisms present,” ≥90% specificity for negatives, and <60 s latency per slide to enable rapid triage. Handoff: Positive or low-confidence slides (e.g., high uncertainty or out-of-distribution alert) are queued to a scientist for confirmation; high-confidence negatives are batch-released for secondary review later in the shift. Quality assurance: Daily control slides are auto-scored; weekly audits trend sensitivity/specificity and review false-negatives. Post-analytical: Structured outputs (Gram call + rationale + ROI thumbnails) are appended to the LIS report with timestamps and versioned model IDs for traceability. Impact: The triage step reduces time-to-first-read for positives, cuts after-hours workload, and standardizes preliminary descriptions while keeping final sign-out with human experts.
VLMs trained on labeled Gram-stained images have shown the ability to produce structured descriptions that encapsulate bacterial morphology, organization, and staining properties. In controlled testing situations, many models attained classification accuracies of 90% in distinguishing common bacterial morphotypes. Nonetheless, difficulties remain in generalizing these models due to varied staining quality, unusual organisms, and imaging discrepancies, particularly in low-resource environments [1].
In addition to stain interpretation, colony morphology represents another domain where VLMs could improve reproducibility. Descriptors, including margin definition, pigmentation, and hemolysis patterns, frequently recorded inconsistently in manual processes, can be systematically collected by models trained on annotated culture photos. However, significant intra-species heterogeneity and a scarcity of available datasets continue to pose technical challenges. Current research has not definitively established whether these algorithms can surpass expert microbiologists in atypical scenarios or in the presence of mixed illnesses.
VLMs provide the capability to automate zone diameter measurements and interpret breakpoints in accordance with guidelines for antimicrobial susceptibility testing (AST). Initial tests indicate over 85% agreement with human annotations in disc diffusion testing; however, performance deteriorates in cases of overlapping zones, indistinct edges, or uncommon infections [8]. Moreover, regulatory and process integration obstacles continue to pose substantial impediments to practical implementation. Figure 1 presents an organized diagram depicting the workflow from slide digitization to VLM-assisted interpretation and LIS integration, while Table 1 summarizes the comparative functionalities of VLMs against traditional methods across several areas.
Haematology
The study of peripheral blood smears is an essential diagnostic method in hematology for assessing erythrocyte morphology, leukocyte differentials, and platelet conditions. Manual review, while helpful, exhibits significant inter-observer variability and is susceptible to reader fatigue. VLMs trained on annotated smears have shown significant improvements in accuracy and interpretability [9].
A study examining six RBC morphologies showed that a vision-language architecture model attained a classification accuracy of 93.4% and an F1-score of 0.91 for sickle cell detection, metrics comparable to those of experienced hematologists. Significantly, VLMs extend beyond mere labeling by generating descriptive narratives, such as identifying hypersegmented neutrophils and associating them with macrocytic anemia patterns. These outputs improve traceability and diminish ambiguity in diagnostic communication [10].
Leukemic pathology adds additional intricacy. Blast detection, Auer rod identification, and left-shifted granulopoiesis recognition necessitate the integration of morphological context with diagnostic reasoning. Certain VLMs, when presented with expert-annotated pictures, have demonstrated the capacity to produce indicative interpretations; nonetheless, they necessitate validation via immunophenotyping and professional corroboration. In preliminary evaluations, metrics such as sensitivity and specificity have varied from 85% to 95%, contingent upon the model, cell type, and staining circumstances [11].
Notwithstanding these advancements, difficulties endure. Numerous datasets are constrained in terms of size, class equilibrium, and demographic heterogeneity. Furthermore, the majority of models have not been subjected to external validation in various laboratory environments, constraining their generalisability. The potential for overfitting to particular staining techniques or imaging resolutions also poses issues regarding transferability.
Histopathological Analysis
Histopathology is the most interpretatively challenging area of diagnostic microscopy. The process entails evaluating tissue architecture, cellular morphology, and context-dependent patterns, typically necessitating years of training. VLMs, when utilized on digitized whole-slide photographs, have demonstrated the capability to produce layered descriptions akin to initial diagnostic impressions [12].
For example, instead of categorizing an image merely as "adenocarcinoma," a VLM would articulate "proliferation of atypical glandular structures exhibiting mitotic figures and nuclear pleomorphism," accompanied by a diagnostic recommendation. In assessments of glandular histology, VLMs exhibit accuracy rates of 87–92% when compared to pathologist annotations, with inter-model variability frequently affected by the diversity of the training corpus and the grade of resolution [13].
In the assessment and classification of tasks—such as the allocation of Gleason or Nottingham scores—VLMs have generated structured outputs consistent with reference standards; nonetheless, error rates escalate in marginal situations and in tissue sections affected by inflammatory or necrotic variables. Restricted availability to high-quality, curated datasets for uncommon malignancies diminishes their robustness across the diagnostic spectrum.
Tasks related to biopsy interpretation, such as the identification of lymphovascular invasion, fibrosis, or necrosis, are inadequately investigated due to the complexities of data annotation. Moreover, interpretability in histology is essential; misdiagnosis may result in substantial clinical repercussions. Consequently, even slight advancements in automation necessitate corresponding stringent validation processes, quality assurance systems, and human oversight checks [14].
In an observational study of laboratory trainees, more than 80% deemed VLM-generated reports more useful than static atlas images [15]. Nonetheless, user happiness does not equate to diagnostic validity. Wider implementation must emphasize regulatory supervision, transparency, and bias reduction—especially when models are developed using institution-specific datasets that may not represent the broader diversity of patients.
Table 1. Comparison of Traditional vs Vision-Language Model Methods in Microscopy.
|
In summary, VLMs show strong promise in augmenting microscopy diagnostics, particularly in enhancing interpretability, improving workflow efficiency, and supporting underserved settings. However, enthusiasm must be balanced with caution. Rigorous external validation, performance benchmarking, and integration into regulated diagnostic frameworks are essential to ensure safety, generalizability, and equitable deployment.
Practical Benefits and Contextual Significance of Vision-Language Models in Microscopy
Vision-Language Models (VLMs) provide a novel approach to enhancing interpretability, consistency, and operational efficiency in microscopy-based diagnostics. Their incorporation into laboratory operations is motivated not solely by innovation but by the quantifiable benefits they provide in standardization, explainability, and scalability. This section rigorously analyses these benefits and delineates their practical significance, while also recognizing existing limitations and areas that need further assessment [16].
Standardization and Consistency in Diagnosis
One big problem with classical microscopy is that it relies on people to interpret the results. Inter-observer variability is still a big problem in diagnostic settings like Gramme stain, leukocyte morphology, and histologic grading, even for seasoned professionals. Vision-language models (VLMs) trained on image-text pairs that have been annotated by experts can help reduce this unpredictability by giving outputs that are structured and consistent [17].
VLMs have been able to correctly classify more than 90% of some morphologies in some areas, with F1-scores ranging from 0.85 to 0.93, depending on how hard the task is and how good the dataset is. Still, their results depend on the exact job and context; they may not work as well in rare situations, with atypical presentations, or with specimens that aren't stained well enough [18]. Because of this, VLMs improve the consistency of diagnoses, but they don't completely rid the need for expert validation or replace established quality assurance systems.
Understanding and Making Clear
Most traditional machine learning models, especially convolutional neural networks (CNNs), only give categorical labels or probability ratings, which might make it hard to put things in a clinical perspective. VLMs make it easier to understand by using clinical language to describe things in a way that makes sense. For example, instead of calling a cell a "blast," a VLM might describe its nuclear-to-cytoplasmic ratio, chromatin arrangement, and nucleolar prominence. This would provide a clear explanation similar to a pathologist's report [19].
This output format makes it easier to understand and more open in clinical settings. It also makes it easier to check for mistakes and encourages communication between lab staff, trainees, and doctors. Recent studies show that VLMs are better at speaking clearly, but they may not be as good at making diagnoses when the situation is unclear compared to human specialists. It's best to think of them as decision support tools rather than replacements for human skills [20].
Workflow Efficacy and Response Duration
Manual microscopy is intrinsically time-consuming. The diagnostic procedure may be prolonged, particularly in high-throughput laboratories or those experiencing personnel shortages, due to the time required for slide scanning and findings documentation. VLMs provide operational benefits by automating the analysis of standard specimens. In experimental installations, automated Gramme stain reporting utilizing VLMs decreased reporting times by more than 50%, with more significant enhancements noted in low-resource settings [21].
Nonetheless, the incorporation of VLMs into laboratory information systems and diagnostic reporting frameworks is complex. Concerns with picture preprocessing, system compatibility, and output standardization must be resolved before extensive implementation. The efficacy of VLMs is consequently linked to infrastructure preparedness and regulatory conformity.
Equity and Remote Diagnostic Assistance
A prominent advantage of VLMs is their capacity to facilitate diagnosis in environments with restricted access to expert microscopy. When integrated with digital slide scanners and cloud-based platforms, VLMs can deliver automated analyses in areas deficient in on-site professionals. Equity in diagnostic access relies not alone on algorithmic efficacy but also on internet connectivity, hardware availability, and language localization [22].
Presently, the majority of VLMs have been trained on datasets sourced from high-resource institutions, perhaps failing to encompass the complete range of clinical presentations observed worldwide. Rectifying this disparity necessitates intentional initiatives to mix training data and assess performance across geographically and demographically varied populations.
Standardization of Educational Support and Training
VLMs demonstrate potential as instruments for medical education. Their capacity to emulate expert analysis and elucidate morphological characteristics in natural language renders them advantageous in educational settings. In controlled investigations, trainees exposed to VLM-generated outputs reported enhancements in morphological comprehension and increased confidence in interpretative tasks [23].
However, the educational benefit must be assessed beyond subjective perceptions. Comparative studies assessing objective learning outcomes—such as pre/post-test performance or retention—are necessary to validate their educational efficacy. Furthermore, model explanations must conform to revised diagnostic criteria and refrain from perpetuating obsolete or inaccurate terminology, as noted in several initial implementations.
Although VLMs offer significant improvements to microscopy operations, their advantages must be understood in relation to their constraints. Diagnostic accuracy is extremely particular to tasks, generalisability is a challenge, and regulatory avenues for clinical application are still developing. Future investigations should emphasize prospective trials, external validations, and implementation in practical clinical environments. Only with such proof can the function of VLMs be distinctly defined and judiciously expanded within laboratory medicine [24].
Challenges in Implementation and Ethical Considerations
Notwithstanding the increasing interest in Vision-Language Models (VLMs), their application in clinical diagnostics is still in its infancy, with numerous unresolved technical, infrastructural, and ethical obstacles. Adoption has been inconsistent and largely restricted to experimental or pilot environments. A 2024 global audit of 520 healthcare facilities revealed that merely 14.7% have included generative AI tools into their diagnostic procedures, with less than 5% utilizing multimodal models like VLMs [25]. Most implementations were primarily situated in academic medical centers or AI-centric consortia, frequently bolstered by specialized bioinformatics teams and tailored infrastructure. The translation into wider clinical practice has been obstructed by apprehensions about model robustness, interpretability, medico-legal risks, and the sufficiency of existing regulatory frameworks [26].
Data Quality and Annotation Constraints
High-performance VLMs rely on extensive, meticulously annotated image-text pairs; yet, medical microscopy datasets are constrained, isolated, and inconsistently organized. In contrast to typical computer vision datasets (e.g., ImageNet), microscope images exhibit significant variability in staining techniques, magnification, resolution, and diagnostic classification. The creation of high-quality datasets, such as annotated Gramme stain panels, comprehensive peripheral smear narratives, or histopathology slide captions, generally necessitates laborious annotation by board-certified experts. Furthermore, semantic inconsistency—exemplified by terminological heterogeneity within institutions or inter-observer discrepancies in morphological classifications—exacerbates the challenges of dataset standardization. These problems impede the scalability and generalisability of trained models [27].
Model Generalisability and Clinical Validation
VLMs trained on data from a restricted range of institutions or geographic areas frequently demonstrate inadequate external validity. The shape of erythrocytes may vary among communities due to endemic hemoglobinopathies or nutritional deficits. Histological characteristics may differ based on fixation methods or the anatomical site of the sample. In the absence of stringent cross-institutional benchmarking, models may underperform when used in unfamiliar clinical settings. Although numerous initial validation endeavors are present, such as the multi-center evaluation of the BioGPT-VL dataset on hematology smears (AUROC 0.84 ± 0.03, n = 4 locations), comprehensive, regulatory-grade investigations are still limited [28]. Consequently, model performance must be rigorously evaluated using varied, representative validation sets and made publicly accessible to guarantee clinical relevance.
Interpretability and User Confidence
Despite VLMs generating natural language outputs, their internal reasoning processes are not transparent. The risk of "hallucinations"—believable yet erroneous interpretations—has been recorded in recent assessments. A comparison study conducted by Yang et al. in 2025 revealed that GPT-4V exhibited a misunderstanding rate of 17.5% for unusual haematologic results, in contrast to 7.2% for pathologist consensus [29]. This disparity can undermine clinician confidence, particularly in critical situations. The use of interpretable characteristics, such as attention heatmaps, structured report templates, and uncertainty scores, may enhance transparency and promote safer implementation. End-user feedback mechanisms are crucial for rectifying model drift and enhancing confidence.
Data Privacy, Security, and Regulatory Compliance
Implementing and utilizing VLMs in healthcare environments presents considerable privacy risks. Microscopy pictures, although less innately recognizable than radiological scans, may nonetheless possess embedded metadata or be linked to uncommon diagnoses. Jurisdictions like the EU and the U.S. impose rigorous safeguards under GDPR and HIPAA, respectively. Moreover, cross-border model training, such as through federated learning or cloud-based fine-tuning, presents legal and ethical challenges concerning data sovereignty. Although technology precautions, including de-identification and encrypted pipelines, are progressing, they require enhancement through strong institutional control and compliance auditing. Regulatory bodies, such as the U.S. FDA and the European Medicines Agency, have not yet granted formal approval for any VLM as an independent diagnostic instrument; existing implementations are classified as “assistive” or “investigational,” necessitating human monitoring [16].
Ethical Supervision and Human-Centric Integration
The ethical implementation of VLMs must reconcile the advantages of automation with protections against excessive dependence and skill degradation. Uncritically accepting automated microscopy results poses a risk of disseminating diagnostic errors, particularly in instances of rare diseases, unknown pathogens, or ambiguous morphologies. Clinical integration must emphasize hybrid workflows, wherein VLM outputs aid rather than supplant expert interpretation. Enhancing the capabilities of laboratory personnel, particularly in resource-limited environments, is crucial to guarantee that automation supports human decision-making rather than supplanting it. Moreover, institutional norms must explicitly define accountability in instances of discrepancies between AI-generated results and clinical outcomes [30].
Synopsis and Prognosis
The integration of VLMs into clinical microscopy is promising yet intricate. It is essential to address critical issues—dataset quality, generalisability, explainability, data governance, and ethical oversight—to go from research prototypes to regulated, real-world implementations. Effective execution necessitates interdisciplinary cooperation among clinical pathology, machine learning, health policy, and bioethics. Future endeavors must emphasize worldwide benchmarking initiatives, improvements in model transparency, and user-centric deployment frameworks to guarantee that VLMs promote diagnostic fairness instead of reinforcing current gaps [31].
Future Directions and Opportunities
Adding Vision-Language Models (VLMs) to medical laboratory operations opens up a number of strategic ways to improve both the accuracy of diagnoses and the training of the staff. One of the most useful short-term uses is in education and training. VLMs can be used as dynamic learning tools that let you add notes to microscope pictures in real time, make differential diagnoses, and create interactive Q&A settings that mimic how experts think. In places where resources are limited and access to senior diagnosticians is limited, these models may be useful as scalable training tools. However, their effectiveness in teaching needs to be proven through rigorous evaluations of user performance before and after exposure [21].
Progress depends on having big, diversified, and expert-annotated microscopy datasets that are not just for education. Medical microscopy data is different from typical image-text datasets used in basic VLM training since the staining, resolution, pathology prevalence, and reporting methods vary from one institution to another. To make sure that everyone is included, people from all around the world need to work together on this. Federated learning and privacy-preserving data harmonization are two examples of projects that can help scale up data collection without putting patient privacy at risk [32].
Another important area for potential growth is systems integration. For VLMs to be useful in real-world diagnostic procedures, they need to work with both Laboratory Information Systems (LIS) and Electronic Health Records (EHR). Integration could make it possible to automatically highlight abnormal morphology, provide draft diagnostic narratives, and link patient histories or test findings across different types of data. Early pilot experiments have shown that it is possible to include massive language models in radiology report workflows. Similar frameworks may also work for pathology and microbiology [33].
Finally, future research should focus on explainability, regulatory preparedness, and human-AI collaboration. VLMs need to be made so that they can clearly explain why a diagnosis was made. For example, they may highlight parts of an image that affect the output or measure how sure they are. Adding feedback loops that let people fix or flag outputs will make things safer and more trustworthy. The ultimate goal is not to replace lab workers but to make decision-support systems that make human knowledge even better, especially in places where there are a lot of samples or not enough staff.
Conclusion
Vision-Language Models (VLMs) are a potential way to use computers for microscopy-based diagnostics because they combine extracting visual features with understanding plain language. Their use in microbiology, hematology, and histopathology may make it easier to automate descriptive reporting, sort through anomalous findings, and make sure that interpretative output is always the same. However, the present implementations are still in the testing phase, and clinical use is limited to pilot-scale deployments in institutions with a lot of resources.
There are many technological and institutional problems that need to be solved before it can be successfully integrated into clinical workflows. These include creating standardized, high-quality training datasets, thorough validation across several sites, strong systems for finding and explaining errors, and following changing rules and regulations for data governance. Without these protections, patient safety could be at risk due to things like model overfitting, diagnostic bias, or too much dependence on automation.
Also, the ethical use of VLMs must always have human monitoring, especially in unusual instances with new presentations or rare diseases. Collaboration between clinical labs, AI developers, and regulatory bodies from other fields will be necessary to make sure that VLMs improve, not replace, clinical reasoning. In the end, we should not see them as independent diagnosticians, but as cognitive aids that are part of human-centered diagnostic ecosystems. To find out how useful and reliable VLMs are in everyday clinical practice, further real-world studies will need to be done to look at their performance, how easy they are to understand, and how well they fit into existing workflows.
Acknowledgements
Acknowledgements: The authors acknowledge all others who played a beneficial role in completing this manuscript.
Funding: The research received no specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability: No datasets were generated or analysed during the current study.
Declarations Competing interests: The authors declare no competing interests.
Clinical trial number: Not applicable.
Ethics, Consent to Participate, and Consent to Publish declarations: Not applicable.
CITATION (Vancouver)
REFERENCES
- Tripathi N, Zubair M, Sapra A. Gram Staining. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 [cited 2025 Jul 23]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK562156/
- Kim I, Kang K, Song Y, Kim TJ. Application of Artificial Intelligence in Pathology: Trends and Challenges. Diagnostics (Basel). 2022 Nov 15;12(11):2794. doi: 10.3390/diagnostics12112794
- Tao L, Zhang H, Jing H, Liu Y, Yan D, Wei G, et al. Advancements in Vision–Language Models for Remote Sensing: Datasets, Capabilities, and Enhancement Techniques. Remote Sens (Basel). 2025 Jan;17(1):162. doi: 10.3390/rs17010162
- Imran MT, Shafi I, Ahmad J, Butt MFU, Villar SG, Villena EG, et al. Virtual histopathology methods in medical imaging - a systematic review. BMC Med Imaging. 2024 Nov 26;24(1):318. doi: 10.1186/s12880-024-01163-7
- Hartsock I, Rasool G. Vision-language models for medical report generation and visual question answering: a review. Front Artif Intell. 2024 Nov 19;7:1430984. doi: 10.3389/frai.2024.1430984
- Vaz JM, Balaji S. Convolutional neural networks (CNNs): concepts and applications in pharmacogenomics. Mol Divers. 2021 Aug;25(3):1569-84. doi: 10.1007/s11030-021-10225-3
- Bertram CA, Stathonikos N, Donovan TA, Bartel A, Fuchs-Baumgartinger A, Lipnik K, et al. Validation of digital microscopy: Review of validation methods and sources of bias. Vet Pathol. 2022 Jan;59(1):26-38. doi: 10.1177/03009858211040476
- Gajic I, Kabic J, Kekic D, Jovicevic M, Milenkovic M, Mitic Culafic D, et al. Antimicrobial Susceptibility Testing: A Comprehensive Review of Currently Used Methods. Antibiotics (Basel). 2022 Mar 23;11(4):427. doi: 10.3390/antibiotics11040427
- Adewoyin AS, Nwogoh B. Peripheral blood film - a review. Ann Ib Postgrad Med. 2014 Dec;12(2):71-9.
- S A, Ganesan K, K BB. A novel deep learning approach for sickle cell anemia detection in human RBCs using an improved wrapper-based feature selection technique in microscopic blood smear images. Biomed Tech (Berl). 2023 Apr 25;68(2):175–85. doi: 10.1515/bmt-2022-0200
- Sehgal T, Sharma P. Auer rods and faggot cells: A review of the history, significance, and mimics of two morphological curiosities of enduring relevance. Eur J Haematol. 2023 Jan;110(1):14-23. doi: 10.1111/ejh.13872
- Imran MT, Shafi I, Ahmad J, Butt MFU, Villar SG, Villena EG, et al. Virtual histopathology methods in medical imaging - a systematic review. BMC Med Imaging. 2024 Nov 26;24(1):318. doi: 10.1186/s12880-024-01163-7
- Khan MYA, Bandyopadhyay S, Alrajjal A, Choudhury MSR, Ali-Fehmi R, Shidham VB. Atypical glandular cells (AGC): Cytology of glandular lesions of the uterine cervix. Cytojournal. 2022 Apr 30;19:31. doi: 10.25259/CMAS_03_11_2021
- Timakova A, Ananev V, Fayzullin A, Makarov V, Ivanova E, Shekhter A, et al. Artificial Intelligence Assists in the Detection of Blood Vessels in Whole Slide Images: Practical Benefits for Oncological Pathology. Biomolecules. 2023 Aug 29;13(9):1327. doi: 10.3390/biom13091327
- Ji J, Hou Y, Chen X, Pan Y, Xiang Y. Vision-Language Model for Generating Textual Descriptions From Clinical Images: Model Development and Validation Study. JMIR Form Res. 2024 Feb 8;8:e32690. doi: 10.2196/32690
- Hartsock I, Rasool G. Vision-language models for medical report generation and visual question answering: a review. Front Artif Intell [Internet]. 2024 Nov 19 [cited 2025 Jul 23];7. Available from: https://www.frontiersin.org/journals/artificial-intelligence/articles/10.3389/frai.2024.1430984/full
- Kim H, Hur M, d'Onofrio G, Zini G. Real-World Application of Digital Morphology Analyzers: Practical Issues and Challenges in Clinical Laboratories. Diagnostics (Basel). 2025 Mar 10;15(6):677. doi: 10.3390/diagnostics15060677
- Takita H, Kabata D, Walston SL, et al. A systematic review and meta-analysis of diagnostic performance comparison between generative AI and physicians. npj Digit Med. 2025;8:175. doi: 10.1038/s41746-025-01543-z
- Sarvamangala DR, Kulkarni RV. Convolutional neural networks in medical image understanding: a survey. Evol Intell. 2022;15(1):1-22. doi: 10.1007/s12065-020-00540-3
- Pillay TS. Increasing the Impact and Value of Laboratory Medicine Through Effective and AI-Assisted Communication. EJIFCC. 2025 Feb 28;36(1):12-25.
- Kurz CF, Merzhevich T, Eskofier BM, Kather JN, Gmeiner B. Benchmarking vision-language models for diagnostics in emergency and critical care settings. NPJ Digit Med. 2025 Jul 10;8(1):423. doi: 10.1038/s41746-025-01837-2
- Jahn SW, Plass M, Moinfar F. Digital Pathology: Advantages, Limitations and Emerging Perspectives. J Clin Med. 2020 Nov 18;9(11):3697. doi: 10.3390/jcm9113697
- Mavrych V, Yousef EM, Yaqinuddin A, Bolgova O. Large language models in medical education: a comparative cross-platform evaluation in answering histological questions. Med Educ Online. 2025 Dec;30(1):2534065. doi: 10.1080/10872981.2025.2534065
- Morales S, Engan K, Naranjo V. Artificial intelligence in computational pathology – Challenges and future directions. Digit Signal Process. 2021 Dec;119:103196. doi: 10.1016/j.dsp.2021.103196
- Jandoubi B, Akhloufi MA. Multimodal Artificial Intelligence in Medical Diagnostics. Information. 2025 Jul;16(7):591. doi: 10.3390/info16070591
- Qi Y, Mohamad E, Azlan AA, Zhang C. Utilization of artificial intelligence in clinical practice: A systematic review of China’s experiences. Digit Health. 2025 Jan-Dec;11:20552076251343752. doi: 10.1177/20552076251343752
- Ali M, Benfante V, Basirinia G, Alongi P, Sperandeo A, Quattrocchi A, et al. Applications of Artificial Intelligence, Deep Learning, and Machine Learning to Support the Analysis of Microscopic Images of Cells and Tissues. J Imaging. 2025 Feb;11(2):59. doi: 10.3390/jimaging11020059
- Safdari A, Keshav CS, Mody D, Verma K, Kaushal U, Burra VK, et al. The external validity of machine learning-based prediction scores from hematological parameters of COVID-19: A study using hospital records from Brazil, Italy, and Western Europe. PLoS One. 2025 Feb 4;20(2):e0316467. doi: 10.1371/journal.pone.0316467
- Yang Z, Yao Z, Tasmin M, Vashisht P, Jang WS, Ouyang F, et al. Unveiling GPT-4V’s hidden challenges behind high accuracy on USMLE questions: Observational Study. J Med Internet Res. 2025 Feb 7;27(1):e65146. doi: 10.2196/65146
- Mennella C, Maniscalco U, De Pietro G, Esposito M. Ethical and regulatory challenges of AI technologies in healthcare: A narrative review. Heliyon. 2024 Feb 29;10(4):e26297. doi: 10.1016/j.heliyon.2024.e26297
- Jung KH. Large Language Models in Medicine: Clinical Applications, Technical Challenges, and Ethical Considerations. Healthc Inform Res. 2025 Apr;31(2):114–124. doi: 10.4258/hir.2025.31.2.114
- Imran MT, Shafi I, Ahmad J, Butt MFU, Villar SG, Villena EG, et al. Virtual histopathology methods in medical imaging - a systematic review. BMC Med Imaging. 2024 Nov 26;24:318. doi: 10.1186/s12880-024-01163-7
- Negro-Calduch E, Azzopardi-Muscat N, Krishnamurthy RS, Novillo-Ortiz D. Technological progress in electronic health record system optimization: Systematic review of systematic literature reviews. Int J Med Inform. 2021 Aug;152:104507. doi: 10.1016/j.ijmedinf.2021.104507
LICENSE



